Rhodopsin is the mammalian dim-light receptor, and the best characterized member of the G protein-coupled receptor (GPCR) family of membrane proteins. We have worked on multiple aspects of rhodopsin function, including the role of lipid-protein interactions in modulating rhodopsin function, the properties of water inside the protein during activation, and the overall mechanism of rhodopsin activation.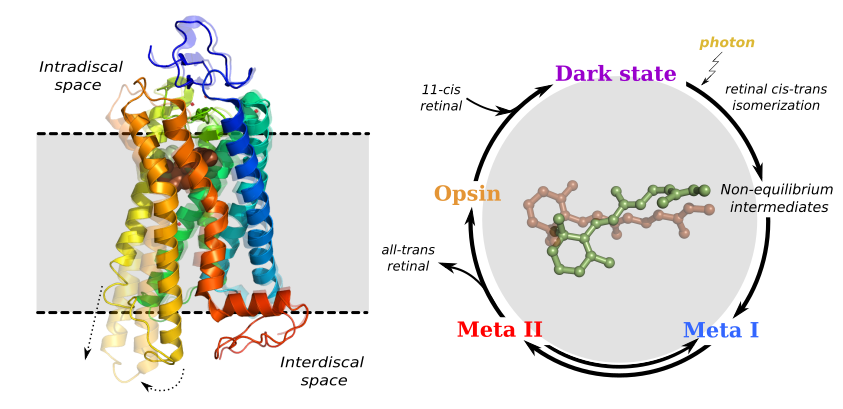
More recently, we’ve been working to model the picosecond-timescale dynamics immediately following photon absorption. This is a collaboration with experimentalists from the University of Arizona (Michael Brown’s lab), Arizona State (Petra Fromme, Rick Kirian, Abhishek Singharoy), and Hauptmann-Woodward (Tom Grant). Using X-ray free-electron lasers, we measure time-resolved small-angle and wide-angle X-ray scattering (SAXS and WAXS) for rhodopsin in detergent micelles. Comparing dark state rhodopsin with activated protein in a pump-probe experiment reveals the perturbations due to photon absorption. We use classical simulations in a similar format to elucidate the atomic-level motions leading to the measured signals.


 e free energy barrier when approaching neutral membranes.
e free energy barrier when approaching neutral membranes.